A Scientific Article By Hasanain.Khaleel.Ibrahim Entitled:Restriction Enzymes:
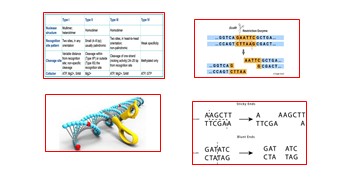
Restriction Enzymes:<br />Werner Arber and Matthew Meselson's laboratory studies of Enterobacteria phage λ (lambda phage) gave rise to the term "restriction enzyme." Restrictions activity were identified by studying the capacity of specific E. coli strains to impede the activity of lambda phage through the enzymatic cleavage of the phage DNA.Because restriction enzymes cleave DNA at or near particular recognition sequences known as restriction sites, they are also referred to as "molecular scissors".<br />The phosphodiester backbone of DNA contains a bond between the phosphorus and 3'-oxygen atoms that is hydrolyzed by restriction enzymes. Mg2+ or other divalent ions are necessary for the enzymes to function.<br />The names of the enzymes begin with an italicized three-letter acronym, like Hind III was the third of four enzymes isolated from Haemophilus influenza serotype d, The first letter denotes the first letter of the genus of bacteria that the enzyme was isolated from, and the next two letters are the species of bacteria that the enzyme was isolated from. To indicate the strain or serotype, these could be followed by additional letters or numbers.<br />The restriction endonucleases are classified into four groups, Type I, II, III, and IV Based on the composition, characteristics of the cleavage site, and the cofactor requirements.<br />Traditionally, restriction enzymes have been classified as I, II, III, and IV. These types differ mainly in their structure, specificity, cleavage site, and cofactors. Two types of ends are produced during restriction digestion of double-stranded DNA: blunt ends and sticky ends.<br /><br /><br /><br /> <br /><br /><br /><br /><br /><br /><br /><br /><br />