Importance of Metabolomics Technology in Advanced Physiology
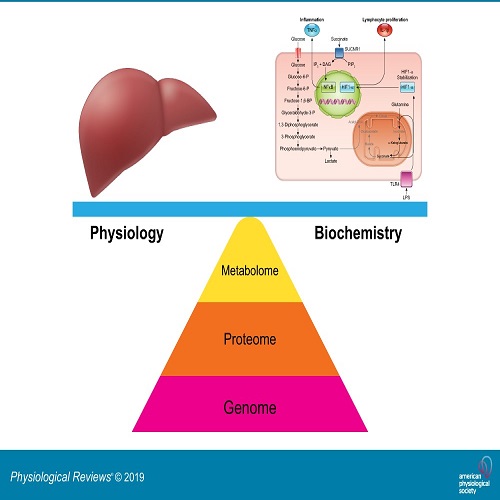
Metabolomics is defined as the identification of all small metabolites (<1000 Da) at a cellular level of any given tissue (the metabolome). These metabolites are comprised of primary compounds, intermediate compounds in metabolic pathways and the end products of cellular processes (Boccard et al., 2010; Nicholson et al., 1999).<br />The first use of metabolomics in the analysis of cellular processes was to determine hundreds of molecules related to redox- state in rat mitochondria (Kristal et al., 1998). Since that time, considerable attention has been given to this area and hundreds of papers were later published about metabolomics (reviewed in Kell, 2004) Metabolomics is commonly used to monitor progression of a disease and the response of therapeutic interventions (Gates & Sweeley, 1978) with a view to potentially identifying early biomarkers of disease or identification of markers of disease progression. However, one of the most important applications of using metabolomics technology is in systems biology (Jewett et al., 2006; Wang et al., 2006). Systems biology is the quantitative study of the whole organism by looking at it as a web of the interactions between molecular components such as DNA, mRNA, proteins, and<br />metabolites with its environment (Nielsen & Jewett, 2007). Constructing systems biology networks is therefore considered to be a novel tool to investigate phenotypic changes and the interaction between phenotype and genotype (Allen et al., 2003; Raamsdonk et al., 2009; Nicholson & Wilson, 2003). Metabolome analysis can be achieved by two different approaches. Firstly, the metabolite profile method (non- targeted metabolomics), in which a large number of metabolites (> 1000 metabolites) are detected. When these are comprised of intracellular metabolites that are commonly known as a metabolomic fingerprint (endometabolome) and extracellular metabolites being called a metabolomic footprint (exometabolome) ((Goodacre et al.,<br />, 2004; Kell, 2004). <br />\<br />The second approach used in metabolomics is a targeted method, through which a relatively small number (<10 metabolites) of specific metabolites can be absolutely measured. However, the number of metabolites detected by the target method has been recently expanded due to advancements in technology (Nielsen & Jewett, 2007; Soltow et al., 2010; Villas-Bôas et al., 2005).<br /><br /><br /><br /><br /><br /><br /><br />Witten by: <br />Assistant Professor Dr. Aqeel Handil Al Jothery (PhD UK, Physiology)<br />Anesthesia Techniques Department, College of Health and Medical Technologies, Al Mustaqbal University<br />Hilla, Iraq<br /><br /><br />ا.م.د عقيل حنظل طارش<br /><br /><br />References:<br /><br />Allen, J., Davey, H. M., Broadhurst, D., Heald, J. K., Rowland, J. J., Oliver, S. G., & Kell, D. B. (2003). High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nature Biotechnology, 21, 692–696. http://doi.org/10.1038/nbt823<br />Boccard, J., Veuthey, J.-L., & Rudaz, S. (2010). Knowledge discovery in metabolomics: an overview of MS data handling. Journal of Separation Science, 33, 290–304. http://doi.org/10.1002/jssc.200900609<br />Gates, S. C., & Sweeley, C. C. (1978). Quantitative metabolic profiling based on gas chromatography. Clinical Chemistry, 24, 1663–1673. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/359193<br />Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G., & Kell, D. B. (2004). Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in Biotechnology, 22, 245–252. http://doi.org/10.1016/j.tibtech.2004.03.007<br />Jewett, M. C., Hofmann, G., & Nielsen, J. (2006). Fungal metabolite analysis in<br />genomics and phenomics. Current Opinion in Biotechnology, 17, 191–197. http://doi.org/10.1016/j.copbio.2006.02.001<br />Kell, D. B. (2004). Metabolomics and systems biology: making sense of the soup.<br /><br />Current Opinion in Microbiology, 7, 296–307. http://doi.org/10.1016/j.mib.2004.04.012<br />Kristal, B. S., Vigneau-Callahan, K. E., & Matson, W. R. (1998). Simultaneous analysis of the majority of low-molecular-weight, redox-active compounds from mitochondria. Analytical Biochemistry, 263, 18–25. http://doi.org/10.1006/abio.1998.2831 <br /><br />Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data.<br />Xenobiotica, 29, 1181–1819. http://doi.org/10.1080/004982599238047<br /><br />Nicholson, J. K., & Wilson, I. D. (2003). Opinion: understanding “global” systems biology: metabonomics and the continuum of metabolism. Nature Reviews. Drug Discovery, 2, 668–676. http://doi.org/10.1038/nrd1157<br />Nielsen, J., & Jewett, M. C. (2007). Metabolomics: a powerful tool in systems biology. Germany: Springer-Verlag Berlin Heidelberg.<br />Raamsdonk, L. M., Teusink, B., Broadhurst, D., Zhang, N., Hayes, A., Walsh, M. C.,<br /> Oliver, S. G. (2009). A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnology, 19, 45– 50. http://doi.org/10.1038/83496<br />Soltow, Q. a, Jones, D. P., & Promislow, D. E. L. (2010). A network perspective on metabolism and aging. Integrative and Comparative Biology, 50, 844–854. http://doi.org/10.1093/icb/icq094<br />Villas-Bôas, S. G., Mas, S., Akesson, M., Smedsgaard, J., & Nielsen, J. (2005). Mass spectrometry in metabolome analysis. Mass Spectrometry Reviews, 24, 613–646. http://doi.org/10.1002/mas.20032<br />Wang, Q.-Z., Wu, C.-Y., Chen, T., Chen, X., & Zhao, X.-M. (2006). Integrating metabolomics into a systems biology framework to exploit metabolic complexity: strategies and applications in microorganisms. Applied Microbiology and Biotechnology, 70, 151–61. http://doi.org/10.1007/s00253-005-0277-2<br /><br />